Search PubMed,
To see number of citations, go to publons.
2025
● Makino, T., Kanada, R., Mori, T., Miyazono, K. I., Komori, Y., Yanagisawa, H., Takada, S., Tanokura, M., Kikkawa, M., Tomishige, M.
Tension-induced suppression of allosteric conformational changes coordinates kinesin-1 stepping.
J Cell Biol 2025
● Hatazawa, S., Fukuda, Y., Kobayashi, Y., Negishi, L., Kikkawa, M., Takizawa, Y., Kurumizaka, H.
Cryo-EM Structures of Native Chromatin Units From Human Cells.
Genes Cells 2025 30:e70019
● An, J., Imasaki, T., Narita, A., Niwa, S., Sasaki, R., Makino, T., Nitta, R., Kikkawa, M.
Dimerization of GAS2 mediates crosslinking of microtubules and F-actin.
EMBO J 2025
2024
● Tani, Y., Yanagisawa, H., Yagi, T., Kikkawa, M.
Structure and function of FAP47 in the central pair apparatus of Chlamydomonas flagella.
Cytoskeleton (Hoboken) 2024 81:669-680
● Morikawa, M., Yamaguchi, H., Kikkawa, M.
Calaxin is a key factor for calcium-dependent waveform control in zebrafish sperm.
Life Sci Alliance 2024
● Goto, S., Tsutsumi, A., Lee, Y., Hosojima, M., Kabasawa, H., Komochi, K., Nagatoishi, S., Takemoto, K., Tsumoto, K., Nishizawa, T., Kikkawa, M., Saito, A.
Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc Natl Acad Sci U S A 2024 121:e2318859121
●Sakuragi, T., Kanai, R., Otani, M., Kikkawa, M., Toyoshima, C., Nagata, S.
The role of the C-terminal tail region as a plug to regulate XKR8 lipid scramblase.
J Biol Chem 2024 :105755
2023
●Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane.
Yanagisawa, H., Kita, Y., Oda, T., Kikkawa, M.
Commun Biol 2023 6:1018
●Kikkawa M.
A quest to unravel the role of the stalk and microtubule-binding domain of axonemal dyneins.
FEBS Lett 2023
●Bui, H. B., Watanabe, S., Nomura, N., Liu, K., Uemura, T., Inoue, M., Tsutsumi, A., Fujita, H., Kinoshita, K., Kato, Y., Iwata, S., Kikkawa, M., Inaba, K.
Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn(2+) uptake into the Golgi apparatus.
Nat Commun 2023 14:4770
●Kubo, T., Tani, Y., Yanagisawa, H. A., Kikkawa, M., Oda, T.
alpha- and beta-tubulin C-terminal tails with distinct modifications are crucial for ciliary motility and assembly.
J Cell Sci 2023
●Shen, H. K., Morishita, K., Hashim, P. K., Okuro, K., Kashiwagi, D., Kimura, A., Yanagisawa, H., Kikkawa, M., Niwa, T., Taguchi, H., Aida, T.
ATP-Responsive Nanoparticles Covered with Biomolecular Machine "Chaperonin GroEL".
Angew Chem Int Ed Engl 2023 :e202304894
●Yamaguchi, H., Morikawa, M., Kikkawa, M.
Calaxin stabilizes the docking of outer arm dyneins onto ciliary doublet microtubule in vertebrates.
eLife 2023 12:e84860
●Malawska, K. J., Takano, S., Oisaki, K., Yanagisawa, H., Kikkawa, M., Tsukuda, T., Kanai, M.
Bioconjugation of Au(25) Nanocluster to Monoclonal Antibody at Tryptophan.
Bioconjug Chem 2023 :
●Chen, Z., Watanabe, S., Hashida, H., Inoue, M., Daigaku, Y., Kikkawa, M., Inaba, K.
Cryo-EM structures of human SPCA1a reveal the mechanism of Ca(2+)/Mn(2+) transport into the Golgi apparatus.
Sci Adv 2023 9:eadd9742
● Ito, C., Makino, T., Mutoh, T., Kikkawa, M., Toshimori, K.
The association of ODF4 with AK1 and AK2 in mice is essential for fertility through its contribution to flagellar shape.
Sci Rep 2023 13:2969
●Takeda, H., Busto, J. V., Lindau, C., Tsutsumi, A., Tomii, K., Imai, K., Yamamori, Y., Hirokawa, T., Motono, C., Ganesan, I., Wenz, L. S., Becker, T., Kikkawa, M., Pfanner, N., Wiedemann, N., Endo, T.
A multipoint guidance mechanism for beta-barrel folding on the SAM complex.
Nat Struct Mol Biol 2023 30:176-187
●Eisenstein, F., Yanagisawa, H., Kashihara, H., Kikkawa, M., Tsukita, S., Danev, R.
Parallel cryo electron tomography on in situ lamellae.
Nat Methods 2023 20:131-138
2022
●Zhang, Y., Kobayashi, C., Cai, X., Watanabe, S., Tsutsumi, A., Kikkawa, M., Sugita, Y., Inaba, K.
Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep 2022 41:111760
●Kobayashi, T., Tsutsumi, A., Kurebayashi, N., Saito, K., Kodama, M., Sakurai, T., Kikkawa, M., Murayama, T., Ogawa, H.
Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun 2022 13:2821
●Kikkawa, M., Yanagisawa, H.
Identifying proteins in the cell by tagging techniques for cryo-electron microscopy.
Microscopy (Oxf) 2022 71:i60-i65
2021
●Nakamura, H., Kikkawa, M., Murata, T.
Technical development and sharing of high-resolution cryo-electron microscopes.
Biophys Physicobiol 2021 18:265-266
●Sakuragi, T., Kanai, R., Tsutsumi, A., Narita, H., Onishi, E., Nishino, K., Miyazaki, T., Baba, T., Kosako, H., Nakagawa, A., Kikkawa, M., Toyoshima, C., Nagata, S.
The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat Struct Mol Biol 2021 28:825-834
●Obata, J., Kawakami, N., Tsutsumi, A., Nasu, E., Miyamoto, K., Kikkawa, M., Arai, R.
Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem Commun (Camb) 2021 57:10226-10229
●Zhang, Y., Watanabe, S., Tsutsumi, A., Kadokura, H., Kikkawa, M., Inaba, K.
Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca(2+) -ATPase SERCA2b.
EMBO J 40:e108482 2021
● Danev, R., Yanagisawa, H., Kikkawa, M.
Cryo-EM Performance Testing of Hardware and Data Acquisition Strategies.
Microscopy (Oxf) 6273016 2021
● Ueda, M., Aoki, T., Akiyama, T., Nakamuro, T., Yamashita, K., Yanagisawa, H., Nureki, O., Kikkawa, M., Nakamura, E., Aida, T., Itoh, Y.
Alternating Heterochiral Supramolecular Copolymerization.
J Am Chem Soc 143:2822-2828 2021
● Sekine, R., Ravat, P., Yanagisawa, H., Liu, C., Kikkawa, M., Harano, K., Nakamura, E.
Nano- and Microspheres Containing Inorganic and Biological Nanoparticles: Self-Assembly and Electron Tomographic Analysis.
J Am Chem Soc 2021 143:2822-2828
● Takeda, H., Tsutsumi, A., Nishizawa, T., Lindau, C., Busto, J. V., Wenz, L. S., Ellenrieder, L., Imai, K., Straub, S. P., Mossmann, W., Qiu, J., Yamamori, Y., Tomii, K., Suzuki, J., Murata, T., Ogasawara, S., Nureki, O., Becker, T., Pfanner, N., Wiedemann, N., Kikkawa, M., Endo, T.
Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature 590:163-169 2021
2020
● Matoba, K., Kotani, T., Tsutsumi, A., Tsuji, T., Mori, T., Noshiro, D., Sugita, Y., Nomura, N., Iwata, S., Ohsumi, Y., Fujimoto, T., Nakatogawa, H., Kikkawa, M., Noda, N. N.
Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat Struct Mol Biol 10.1038/s41594-020-00518-w 2020
● Lu, H., Nakamuro, T., Yamashita, K., Yanagisawa, H., Nureki, O., Kikkawa, M., Gao, H., Tian, J., Shang, R., Nakamura, E.
B/N-Doped p-Arylenevinylene Chromophores: Synthesis, Properties, and Microcrystal Electron Crystallographic Study.
J Am Chem Soc 142:18990-18996 2020
● Tojo, S., Zhang, Z., Matsui, H., Tahara, M., Ikeguchi, M., Kochi, M., Kamada, M., Shigematsu, H., Tsutsumi, A., Adachi, N., Shibata, T., Yamamoto, M., Kikkawa, M., Senda, T., Isobe, Y., Ohto, U., Shimizu, T.
Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun 11:5204 2020
● Zhang, Y., Inoue, M., Tsutsumi, A., Watanabe, S., Nishizawa, T., Nagata, K., Kikkawa, M., Inaba, K.
Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv 6:eabb0147 2020
● Nakamura, R., Numata, T., Kasuya, G., Yokoyama, T., Nishizawa, T., Kusakizako, T., Kato, T., Hagino, T., Dohmae, N., Inoue, M., Watanabe, K., Ichijo, H., Kikkawa, M., Shirouzu, M., Jentsch, T. J., Ishitani, R., Okada, Y., Nureki, O.
Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol 3:240 2020
●Hiroyoshi Hamada ,Takayuki Nakamuro ,Keitaro Yamashita ,Haruaki Yanagisawa ,Osamu Nureki ,Masahide Kikkawa ,Koji Harano ,Rui Shang, and Eiichi Nakamura
Spiro-conjugated Carbon/Heteroatom-bridged p-Phenylenevinylenes: Synthesis, Properties, and Microcrystal Electron Crystallographic Analysis of Racemic Solid Solutions
Bulletin of the Chemical Society of Japan 93:776, 2020
● Nishida, N., Komori Y., Takarada O., Watanabe A., Tamura S., Kubo S., Shimada I. & Kikkawa M.
Structural basis for two-way communication between dynein and microtubules
Nat Commun. 11:1038, 2020
2019
● Araiso, Y., Tsutsumi, A., Qiu, J., Imai, K., Shiota, T., Song, J., Lindau, C., Wenz, L. S., Sakaue, H., Yunoki, K., Kawano, S., Suzuki, J., Wischnewski, M., Schutze, C., Ariyama, H., Ando, T., Becker, T., Lithgow, T., Wiedemann, N., Pfanner, N., Kikkawa, M., Endo, T.
Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature 575:395-401, 2019
● Ito, C., Akutsu, H., Yao, R., Yoshida, K., Yamatoya, K., Mutoh, T., Makino, T., Aoyama, K., Ishikawa, H., Kunimoto, K., Tsukita, S., Noda, T., Kikkawa, M., Toshimori, K.
Odf2 haploinsufficiency causes a new type of decapitated and decaudated spermatozoa, Odf2-DDS, in mice.
Sci Rep 9:14249, 2019
●Keita Sasaki, Kogiku Shiba, Akihiro Nakamura, Natsuko Kawano, Yuhkoh Satouh, Hiroshi Yamaguchi, Motohiro Morikawa, Daisuke Shibata, Ryuji Yanase, Kei Jokura, Mami Nomura, Mami Miyado, Shuji Takada, Hironori Ueno, Shigenori Nonaka, Tadashi Baba, Masahito Ikawa, Masahide Kikkawa, Kenji Miyado & Kazuo Inaba
Calaxin is required for cilia-driven determination of vertebrate laterality
Communications Biology 2:226, 2019
●Danev, R., Yanagisawa, H., Kikkawa, M.
Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions.
Trends Biochem Sci 44:837-848, 2019
● Owa M, Uchihashi T., Yanagisawa HA, Yamano T., Iguchi H., Fukuzawa H., Wakabayashi KI, Ando T., Kikkawa M.
Inner lumen proteins stabilize doublet microtubules in cilia and flagella.
Nat Commun. 10:1143, 2019
2018
●Hiramoto, E., Tsutsumi, A., Suzuki, R., Matsuoka, S., Arai, S., Kikkawa, M., Miyazaki, T.
The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein.
Science Advances, 4:eaau1199, 2018
● Kasuya, G., Nakane, T., Yokoyama, T., Jia, Y., Inoue, M., Watanabe, K., Nakamura, R., Nishizawa, T., Kusakizako, T., Tsutsumi, A., Yanagisawa, H., Dohmae, N., Hattori, M., Ichijo, H., Yan, Z., Kikkawa, M., Shirouzu, M., Ishitani, R., Nureki, O.
Cryo-EM structures of the human volume-regulated anion channel LRRC8
Nat Struct Mol Biol. 25(9):797-804, 2018
● Yamaguchi H., T. Oda, M. Kikkawa, H. Takeda
Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly
eLife 2018;7:e36979
● Koshimizu S., Kofuji R., Sasaki-Sekimoto Y., Kikkawa M., Shimojima M., Ohta H., Shigenobu S., Kabeya Y., Hiwatashi Y., Tamada Y., Murata T., Hasebe M.
Physcomitrella MADS-box genes regulate water supply and sperm movement for fertilization
Nature Plants 4, 36-45, 2018
2017
●Kuroki K., K. Mio, A. Takahashi, H. Matsubara, Y. Kasai, S. Manaka, M. Kikkawa, D. Hamada, C. Sato, K. Maenaka
Class II-like Structural Features and Strong Receptor Binding of the Nonclassical HLA-G2 Isoform Homodimer.
J Immunology 198:3399, 2017
2016
● Wang D., R. Nitta, M. Morikawa, H. Yajima, S. Inoue, H. Shigematsu, M. Kikkawa , and N. Hirokawa
Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A.
Elife 2016 Sep 30;5. pii: e18101. doi: 10.7554/eLife.18101.
● Yamagishi M., H. Shigematsu, T. Yokoyama, M. Kikkawa, M. Sugawa, M. Aoki, M. Shirouzu, J. Yajima, R. Nitta
Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility.
Structure, 24:1-13, 2016
● Oda T., T. Abe, H. A. Yanagisawa, and M. Kikkawa
Docking complex-independent alignment of outer dynein arms with 24-nm periodicity in vitro.
Journal of Cell Science, 129:1547-51, 2016
● Oda T., T. Abe, H. A. Yanagisawa, and M. Kikkawa
Structure and function of outer dynein arm intermediate and light chain complex
Molecular Biology of the Cell, 27:1051-9, 2016
2015
Oda T., H. A. Yanagisawa, and M. Kikkawa
Detailed Structural and biochemical characterization of the nexin-dynein regulatory complex.
Molecular Biology of the Cell, 26:294-304, 2015
2014
Oda T., H. A. Yanagisawa, R. Kamiya, and M Kikkawa
A molecular ruler determines the repeat length in eukaryotic cilia and flagella
Science, 346:857-860, 2014
Free Access to : Abstract, Reprint, and PDF
Related EMDB: 6109-6117
Fujita S., T. Matsuo, M. Ishiura, M. Kikkawa
High-Throughput Phenotyping of Chlamydomonas Swimming Mutants Based on Nanoscale Video Analysis
Biophysical Journal, 107:336-345, 2014
Yanagisawa HA, G. Mathis, T. Oda, M. Hirono, E. A. Richey, H. Ishikawa, W. F. Marshall, M. Kikkawa, and H. Qin.
FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas.
Molecular Biology of the Cell, 25:1472-1483, 2014
(Cover Photo)

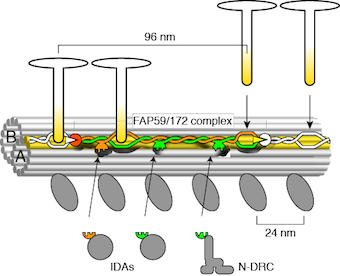
Oda T., HA Yanagisawa, T. Yagi, and M. Kikkawa
Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity
Journal of Cell Biology, 204:807-819, 2014
related EMDB: 5845, 5846, 5847, 5848, 5849, 5850, 5851, 5852, 5853, 5854, 5855
Takarada O., N. Nishida, M. Kikkawa, and I. Shimada
Backbone and side-chain 1H, 15N and 13C resonance assignments of the microtubule-binding domain of yeast cytoplasmic dynein in the high and low-affinity states
Biomolecular NMR Assignments, 8:372-382, 2014
2013
Oda T. and M. Kikkawa
Novel Structural Labeling Method using Cryo-electron Tomography and Biotin-Streptavidin System
Journal of Structural Biology, 183:305-11, 2013
related EMDB: 5711, 5712
Kikkawa M.
Big steps toward understanding dynein
Journal of Cell Biology, 202:15-23, 2013

Oda T., T. Yagi, H. Yanagisawa, and M. Kikkawa
Identification of the Outer-Inner Dynein Linker as a Hub Controller for Axonemal Dynein Activities
Current Biology, 23:656-64, 2013
related EMDB: 5618, 5619, 5620, 5621, 5622
2009
Bodey A. J., M. Kikkawa and C. A. Moores
9Å structure of a microtubule-bound mitotic motor
Journal of Molecular Biology, 388:218-224, 2009
2008
Kikkawa M.
The role of microtubule in processive kinesin movement.
Trends in Cell Biology, 18:128-135 , 2008
2007
Mizuno N., A. Narita, T. Kon, K. Sutoh, and M. Kikkawa
Three-dimensional structure of cytoplasmic dynein bound to microtubules.
Proc. Natl. Acad. Sci. USA, 104:20832-7, 2007

Narita A., N. Mizuno, M. Kikkawa, and Y. Maeda
Molecular determination by electron microscopy of the dynein-microtubule complex structure.
Journal of Molecular Biology, 372:1320-1336, 2007

Oda T., N. Hirokawa, and M. Kikkawa
Three-dimensional structures of flagellar dynein-microtubule complex by cryo-electron microscopy.
Journal of Cell Biology 177:243-252, 2007

Morfini G., G. Pigino, N. Mizuno, M. Kikkawa, and S. T. Brady
Tau binding to microtubules does not directly affect microtubule-based vesicle motility.
J. Neurosci Research 85:2620-2630, 2007

Metlagel, Z., Y. S. Kikkawa, and M. Kikkawa
Ruby-Helix : An implementation of helical image processing based on object-oriented scripting language
Journal of Structural Biology 157:95-105, 2007

2006
Kikkawa, M. and Z. Metlagel
A molecular "zipper" for microtubules. (preview)
Cell, 127(7):1302-4, 2006
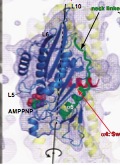
Kikkawa, M. and N. Hirokawa
High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations
EMBO Journal, 25:4187-4194, 2006
Arac, D., X. Chen, H. A. Khant, J. Ubach, S. J. Ludtke, M. Kikkawa, A. E. Johnson, W. Chiu, T. C. Südhof, and J. Rizo
Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids
Nature Structural & Molecular Biology, 13:209-17, 2006

2005
Wu, X., P. B. Alexander, Y. He, M. Kikkawa, P. D. Vogel and S. L. McKnight
Mammalian Sprouty Proteins assemble into large, monodisperse particles having the properties of intracellular Nanobatteries
Proc. Natl. Acad. Sci. U. S. A., 102:14058-62, 2005
2004
Kikkawa, M
A new theory and algorithm for reconstructing helical structures with a seam.
Journal of Molecular Biology, 343:943-955, 2004

Mizuno N., S. Toba, M. Edamatsu, J. Watai-Nishii, N. Hirokawa, Y. Y. Toyoshima and M. Kikkawa
Dynein and kinesin share the same microtubule binding site.
EMBO Journal, 23:2459-2467, 2004
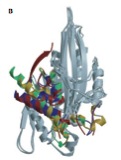
Nitta R., M. Kikkawa, Y. Okada, and N. Hirokawa
Swinging movement of the back door of kinesin generates the alternating use of its two microtubule-binding loops
Science, 305:678-683, 2004
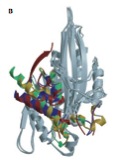
2003
Honma N., Y. Takei, Y. Tanaka, T. Nakata, S. Terada, M. Kikkawa, Y. Noda, and N. Hirokawa
Kinesin Superfamiliy Protein 2A (KIF2A) Functions in Suppression of Collateral Branch Extension
Cell, 114:229-239, 2003
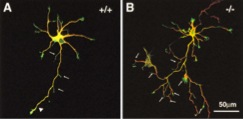
1994-2001
Kikkawa, M., E. P. Sablin, Y. Okada, H. Yajima, RJ Fletterick and N. Hirokawa
Switch Based Mechanism of Kinesin Motors.
Nature, 411:439-445, 2001
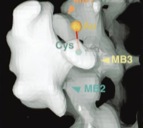
Kikkawa, M., Y. Okada, and N. Hirokawa
15Å Resolution Model of the Monomeric Kinesin Motor, KIF1A.
Cell, 100:241-252, 2000
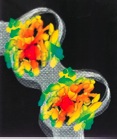
Wolf, S. G., E. Nogales, M. Kikkawa, D. Gratzinger, N. Hirokawa and K. H. Downing
Interpreting a medium-resolution model of tubulin: comparison of Zinc-sheet and microtubule structure.
Journal of Molecular Biology, 262: 485-501, 1996
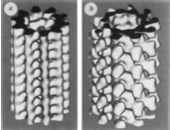
Kikkawa, M., T. Ishikawa, T. Wakabayashi, and N. Hirokawa
Three-dimensional structure of the kinesin head-microtubule complex.
Nature, 376:274- 276. 1995
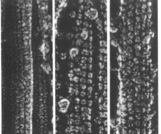
Kikkawa, M., I. Ishikawa, T. Nakata, T. Wakabayashi and N. Hirokawa
Direct visualization of the microtubule lattice seam both in vitro and in vivo.
Journal of Cell Biology, 127: 1965-1971, 1994